HUNK Protein Aggregation Predictor
The routine measurement of protein stability in different formulations using chemical denaturation used to be all but impossible, or at the very least time consuming and tedious. Well not anymore! The fully automated HUNK enables you to perform these experiments with a minimum of fuss providing you with valuable data on whether or not your protein is likely to aggregate over time. Try to do this with any other system.
HUNK will allow you to determine 96 ?Gs on 96 different conditions. The automated operation means all this data will be produced without any operator interaction once the samples have been loaded. This resultant data will help you to choose which of your protein formulations should be taken through to the optimisation phase.
The HUNK allows you to measure:
- ?G measure stability
- ?Gtrend predict aggregation
- KD small molecule affinity
The Importance of ?G
?G lets you know what you have at room temperature. Chemical denaturation is the only way to get to ?G which quantifies the equilibrium between a biologics native and denatured states.
A high ?G value is generally more desirable and means your protein is less denatured. A low ?G value for your protein means it is more denatured and hence, less stable.
HUNK allows you to determine whether your formulation is ready to use or if it needs further work.
Automated Operation
Determination of ?G used to be difficult and time consuming.
These types of measurement are now much more simplified. All you need to do is set the measurement conditions, anywhere from a few to dozens, add your protein samples, formulations and denaturant and set it running. HUNK will then produce your data automatically. HUNK will operate unattended for hours or even days until your experiment has completed.

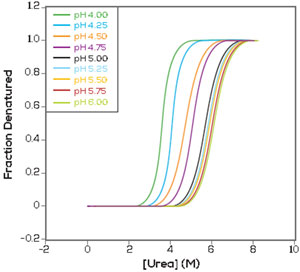 ?G Determination in an Hour
?G Determination in an Hour
HUNK allows you determine ?G in as little as one hour. It does this by building a denaturation curve by increasing the concentration of denaturant until the proteins completely unfold.
C1/2 is the midpoint of the curve, or the concentration of denaturant required to onehalf unfold the protein. HUNK multiplies C1/2 by the slope (m value) of the curve at C1/2 to calculate ?G for your protein.
Regardless of whether you are comparing 2 conditions or 96, you can easily see the effect of your additives (e.g. pH change or small salt addition) on the stability of your biologic. This allows you to choose the optimal conditions to optimise or to perform an aggregation prediction.
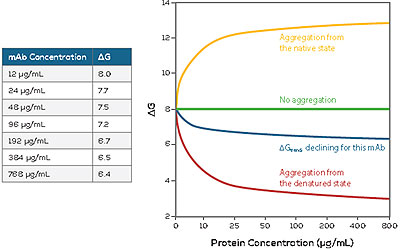 Aggregation Prediction
Aggregation Prediction
HUNK is also able to measure changes in ?G as a function of increasing protein concentration, i.e. ?Gtrend. This provides valuable information on aggregation propensity and pathway.
If your protein does aggregate, HUNK is able to tell you if it happened in the native or denatured state, what fraction is denatured, how much is aggregated and how much the denatured protein aggregates.
Knowing this information early on will help you concentrate on the formulations most likely to succeed, saving you time and effort.







